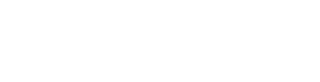RNA silencing in Mucor circinelloides
During the last years, our research group has been involved in the molecular dissection of the RNA silencing mechanism in fungi, using M. circinelloides as a model organism. This regulatory mechanism leads to the suppression of gene expression by small non-coding RNA molecules (sRNAs) of 20 to 30 nucleotides, generated by several different pathways. Our group demonstrated the existence of this mechanism in Mucor some years ago, and since then, it has identified and functionally characterized the key genes involved in the silencing pathway, obtaining mutants in each of them. Several of these mutants are deficient in the vegetative development and other processes such as autolysis induced by nutritional stress, suggesting the involvement of the silencing machinery in the control of endogenous functions. To investigate the endogenous functions of the silencing mechanism in Mucor we used deep sequencing techniques to analyze the repertoire and functional diversity of sRNAs in this organism. This allowed us to identify different classes of sRNAs, which derive from exons and regulate the expression of the protein coding genes from which they were produced. These classes differ in their structural characteristics and proteins involved in their biogenesis, which highlights the complexity of the silencing pathways in fungi. Analysis of the identified sRNAs and their target genes suggests that the silencing machinery could participate, among others, in very important biological processes in Mucor, such as light responses and pathogenesis. Our research now focuses on the connection between specific sRNAs and the Mucor responses to different external and internal stimuli.